overview
These data support: J. J. Gillespie, C. H. McKenna, M. J. Yoder, R. R. Gutell, J. S. Johnston, J. Kathirithamby, A. I. Cognato. 2005. Assessing the odd secondary structural properties of nuclear small subunit ribosomal RNA sequences (18S) of the twisted-wing parasites (Insecta: Strepsiptera). Insect Mol. Biol. 15: 625-643 
abstract
We report the entire sequence (2864 nts) and secondary structure of the nuclear small subunit ribosomal RNA (SSU rRNA) gene (18S) from the twisted-wing parasite Caenocholax fenyesi texensis Kathirithamby & Johnston (Strepsiptera: Myrmecolacidae). The majority of the base pairings in this structural model map on to the SSU rRNA secondary and tertiary helices that were previously predicted with comparative analysis. These regions of the core rRNA were unambiguously aligned across all Arthropoda. In contrast, many of the variable regions, as previously characterized in other insect taxa, had very large insertions in C. f. texensis. The helical base pairs in these regions were predicted with a comparative analysis of a multiple sequence alignment (that contains C. f. texensis and 174 published arthropod 18S rRNA sequences, including eleven strepsipterans) and thermodynamic-based algorithms. Analysis of our structural alignment revealed four unusual insertions in the core rRNA structure that are unique to animal 18S rRNA and in general agreement with previously proposed insertion sites for strepsipterans. One curious result is the presence of a large insertion within a hairpin loop of a highly conserved pseudoknot helix in variable region 4. Despite the extraordinary variability in sequence length and composition, this insertion contains the conserved sequences 5'-AUUGGCUUAAA-3' and 5'-GAC-3' that immediately flank a putative helix at the 5'- and 3'-ends, respectively. The longer sequence has the potential to form a nine base pair helix with a sequence in the variable region 2, consistent with a recent study proposing this tertiary interaction. Our analysis of a larger set of arthropod 18S rRNA sequences has revealed possible errors in some of the previously published strepsipteran 18S rRNA sequences. Thus we find no support for the previously recovered heterogeneity in the 18S molecules of strepsipterans. Our findings lend insight to the evolution of RNA structure and function and the impact large insertions pose on genome size. We also provide a novel alignment template that will improve the phylogenetic placement of the Strepsiptera among other insect taxa.
data
- Online reports are available in the interleave index.
- The present model version is 18S_arthropods.00.04.Nex.
- The present stem index file is 18S_arthropods.SI.00.01.txt.
- There is a working jrna template (requires ver 0.82 in `working` folder) here
- Joe's excel explanation file: Arthropod_18S.xls
- Past version- 18S_arthropods.00.02.Nex (used in paper).
- Past version- 18S_arthropods.00.03.Nex (minor revision).
figures
Use the "view-image" function of your browser (right-click->"view image" (ctrl-click for Mac) in Mozilla/Netscape) or download the images to examine them at full resolution.
Figures are numbered as in Gillespie et al. (Submittted).
figure 1
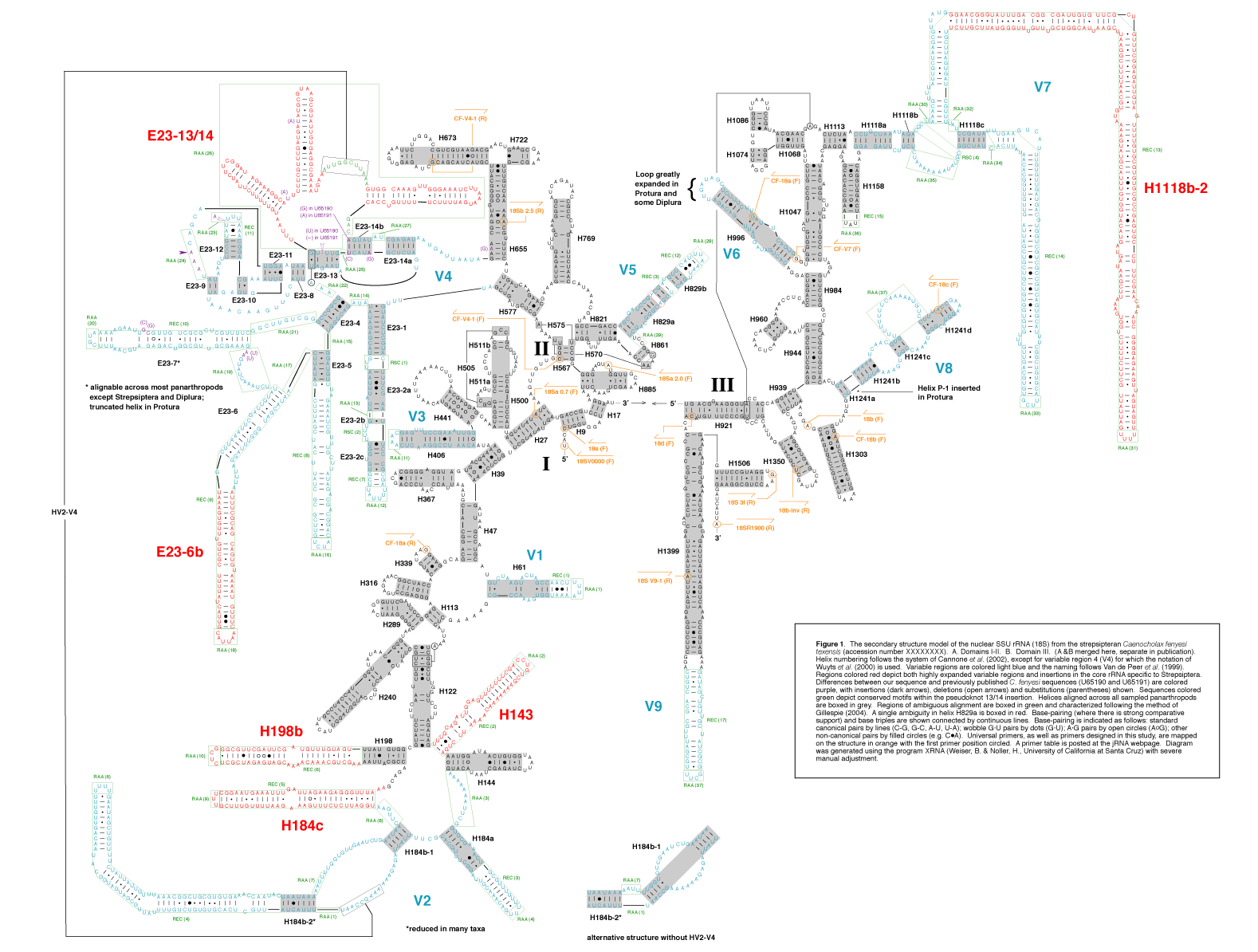
figure 2
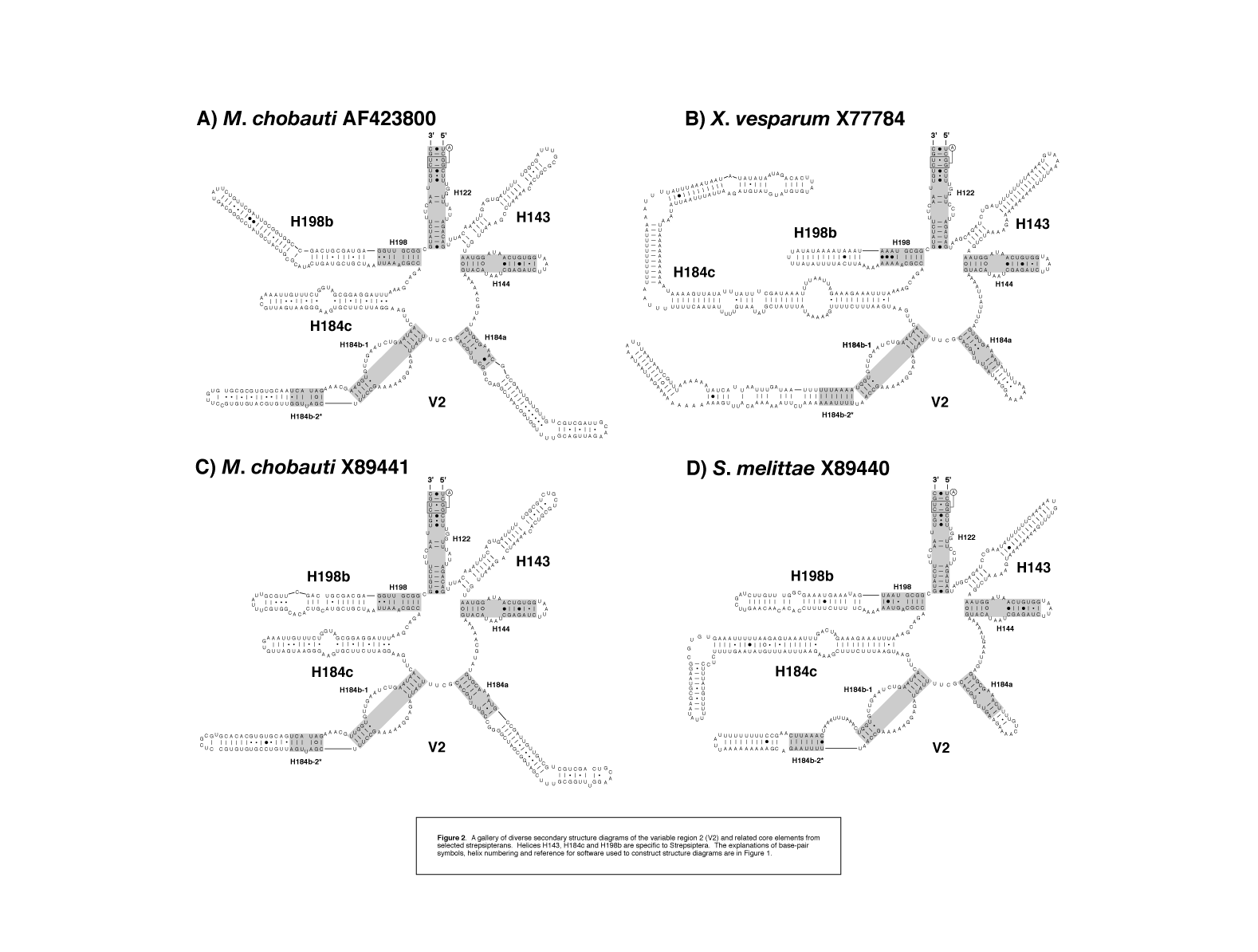
figure 3
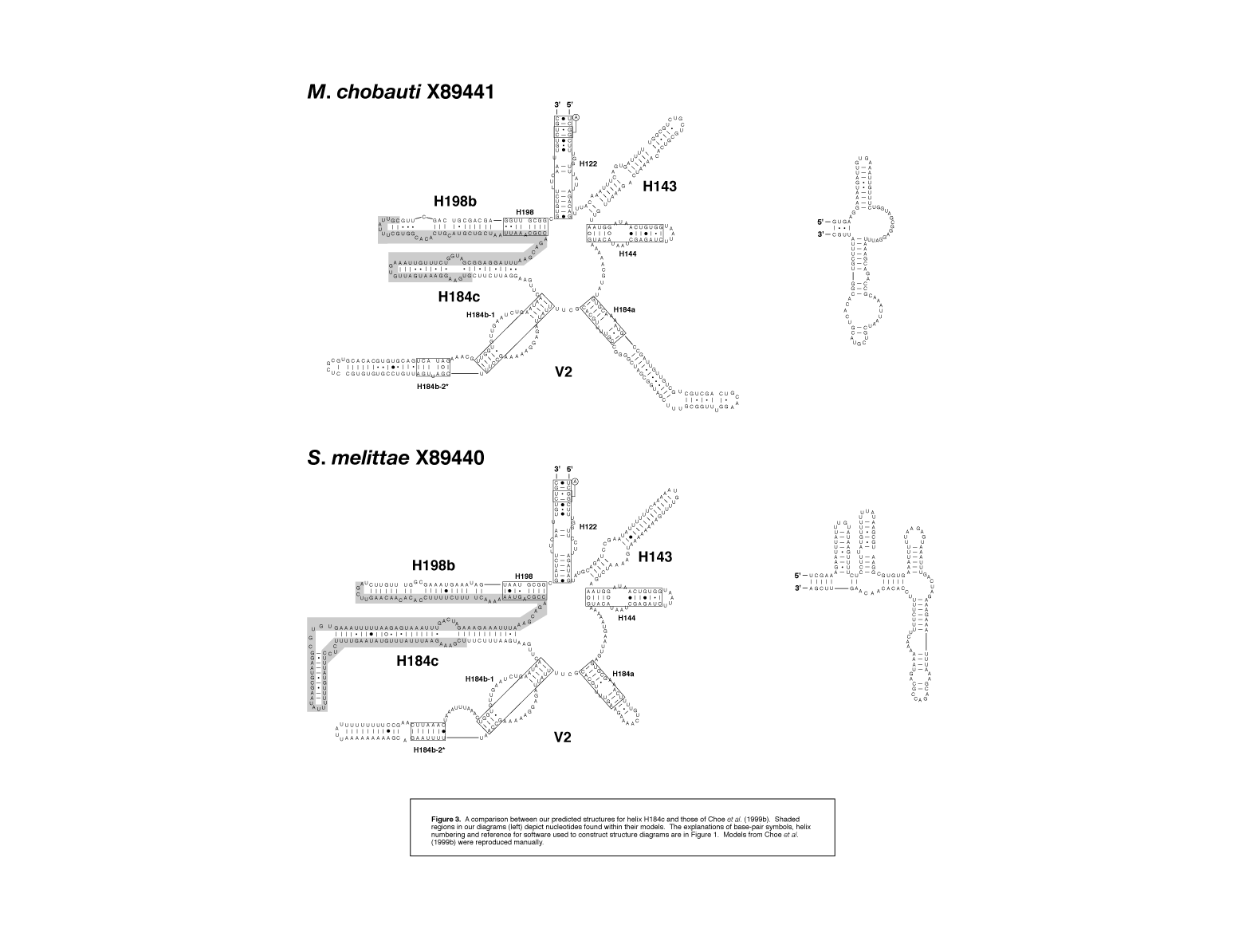
figure 4AC
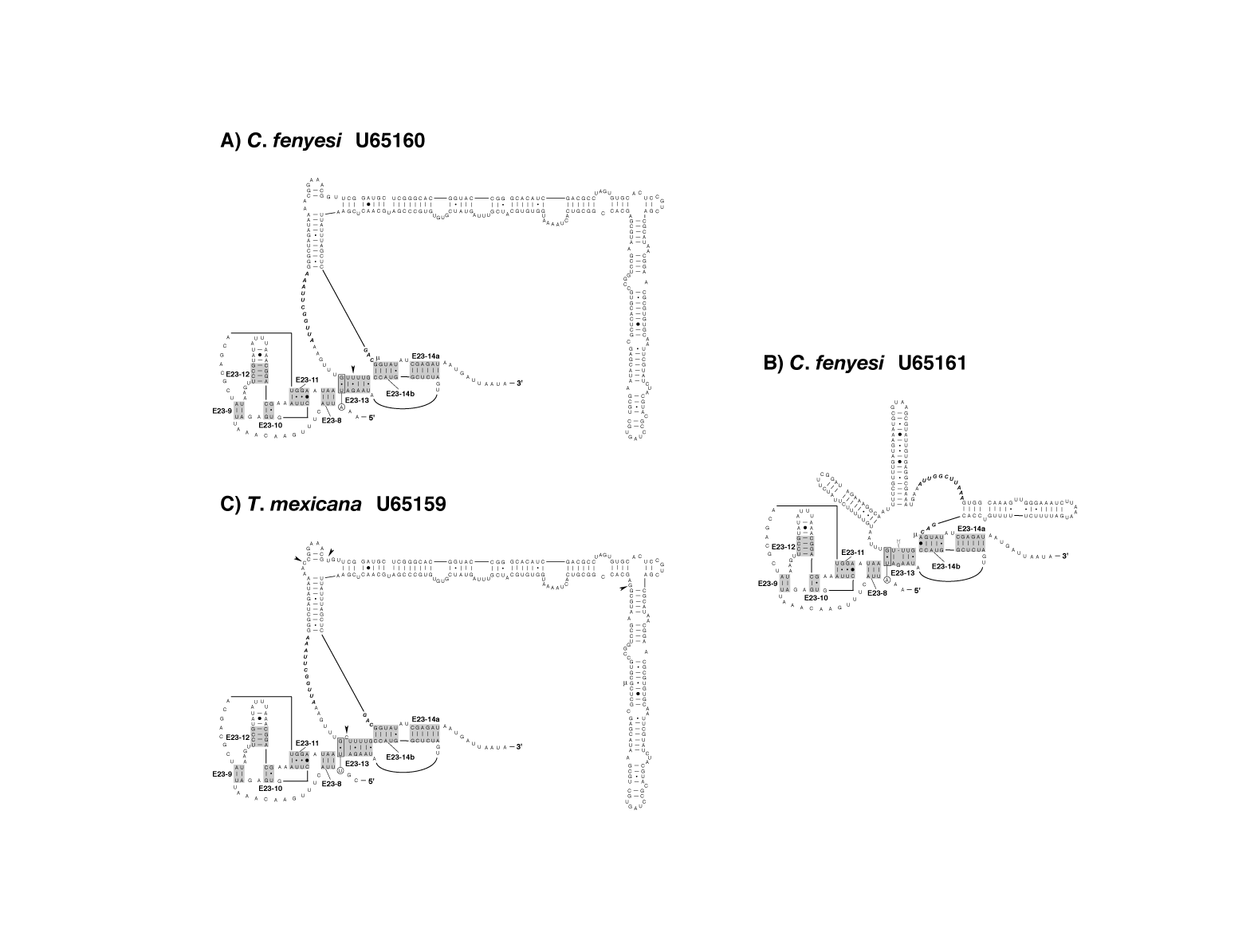
figure 4DE
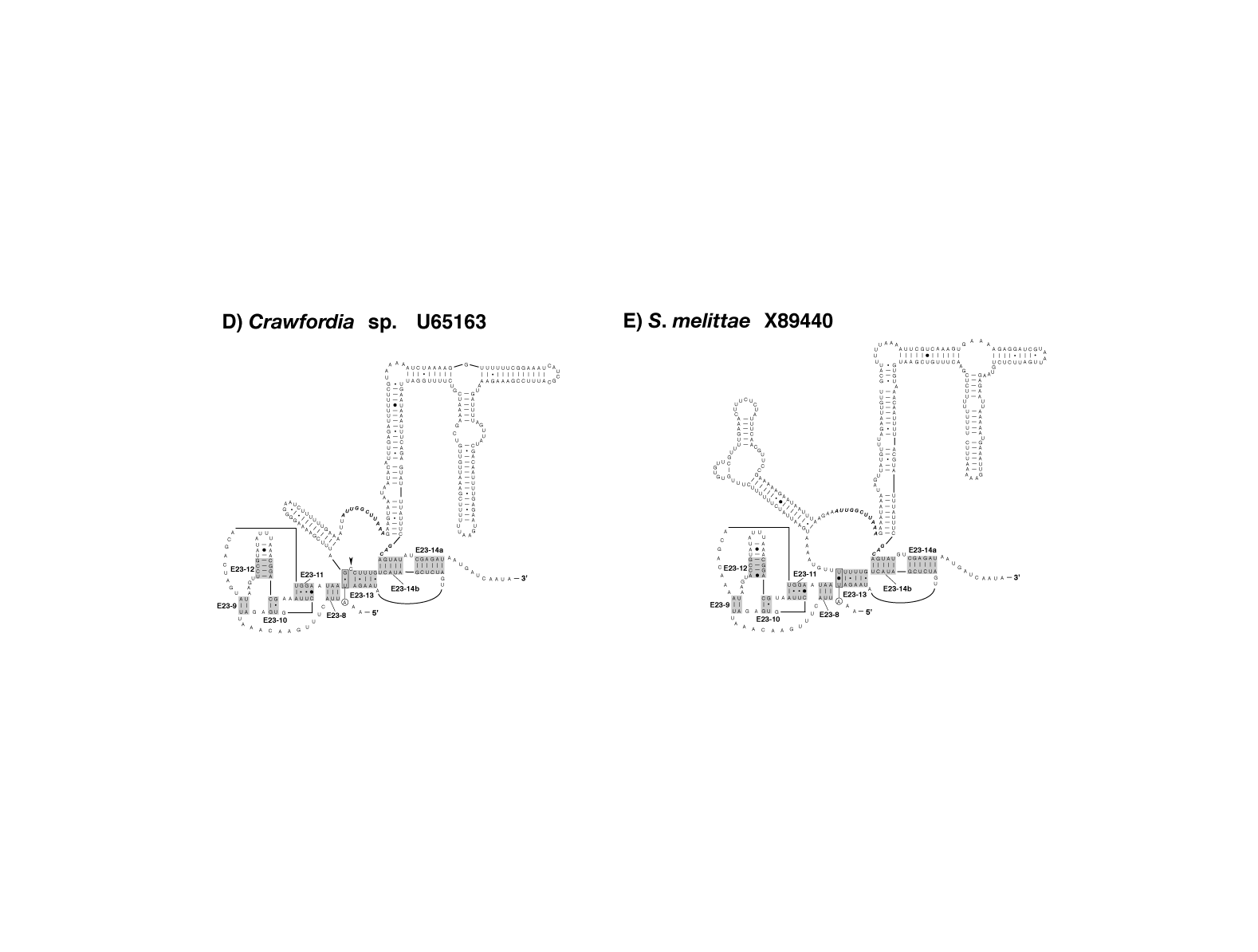
figure 4FH
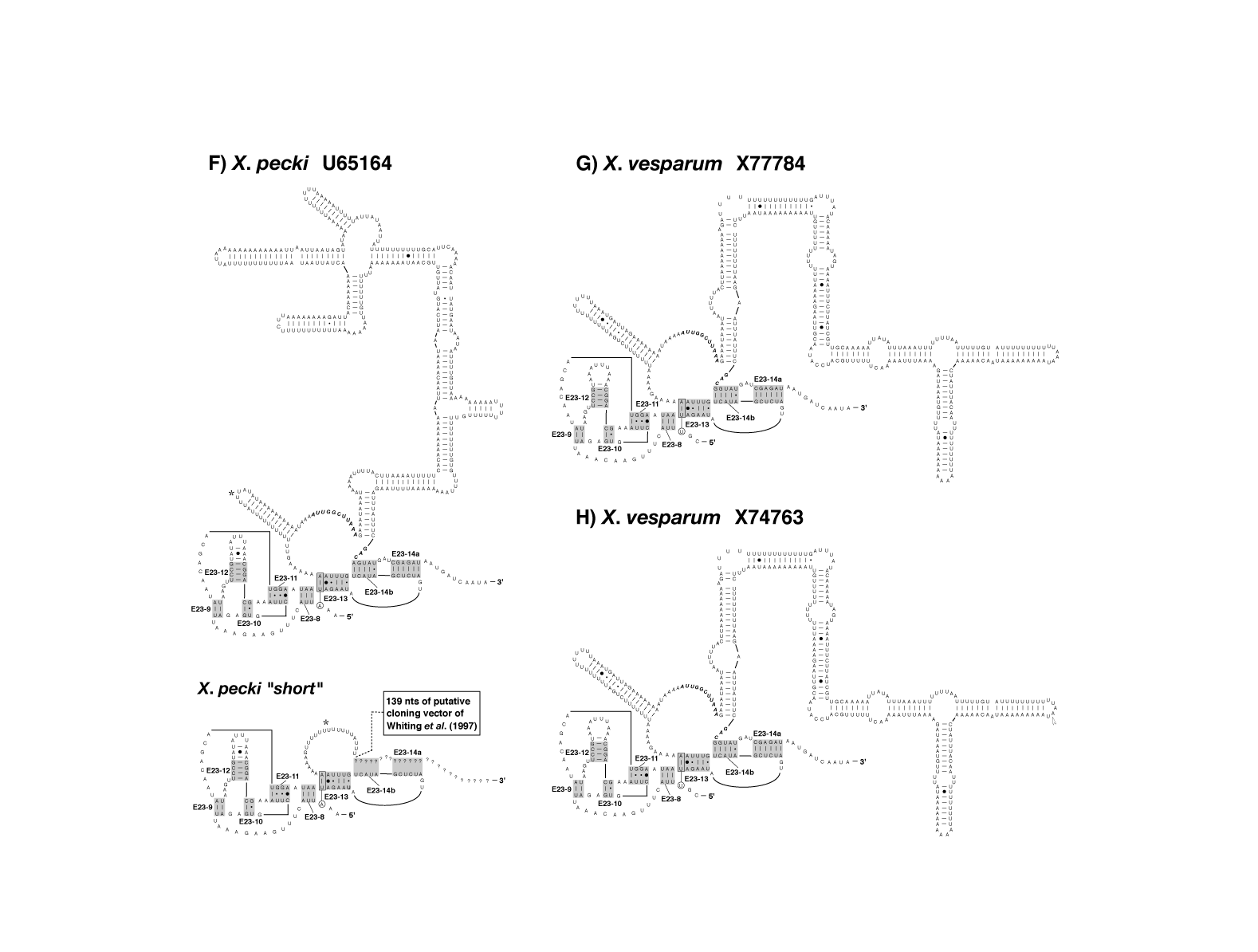
figure 4IJ
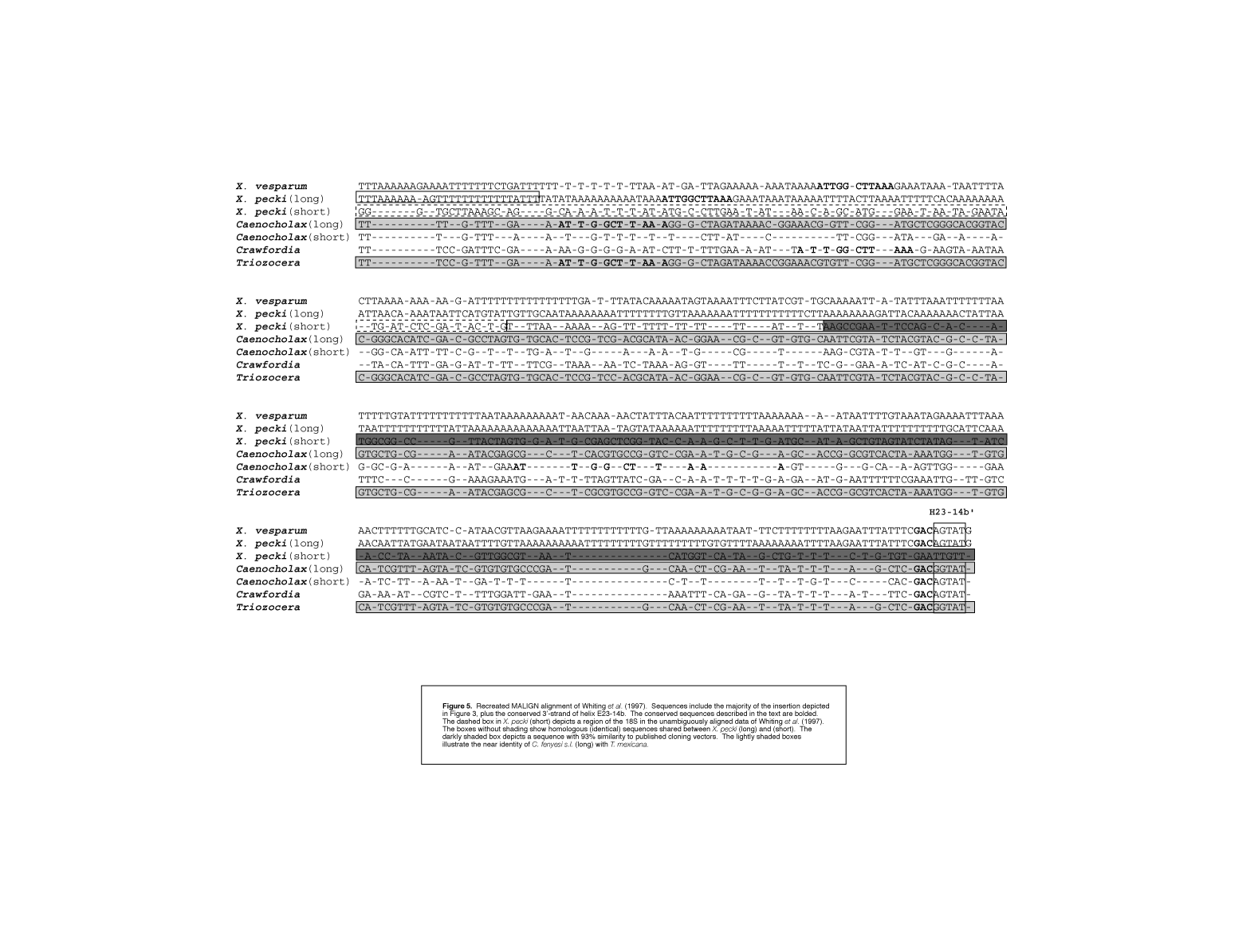
figure 5
tables
table 1
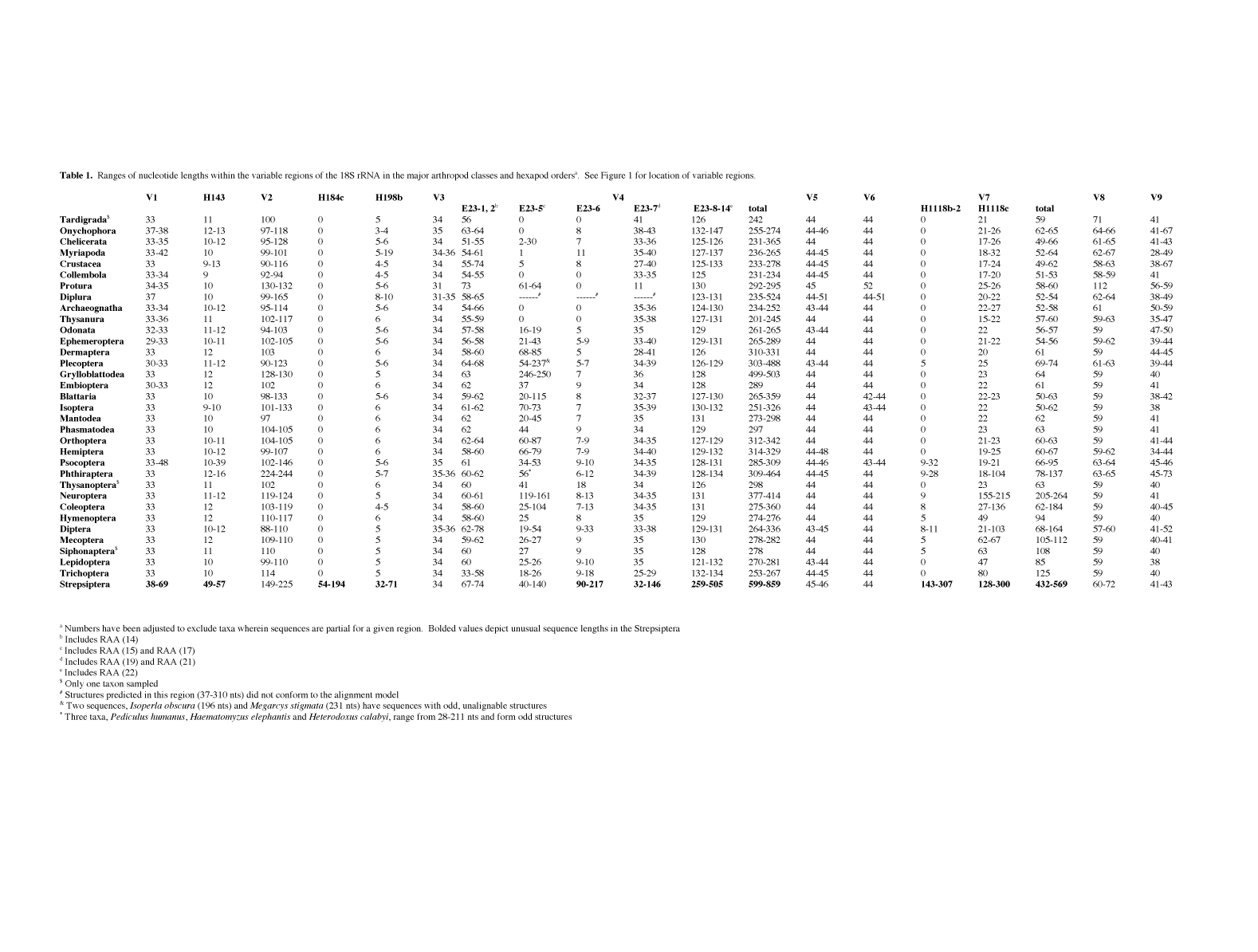
table 2
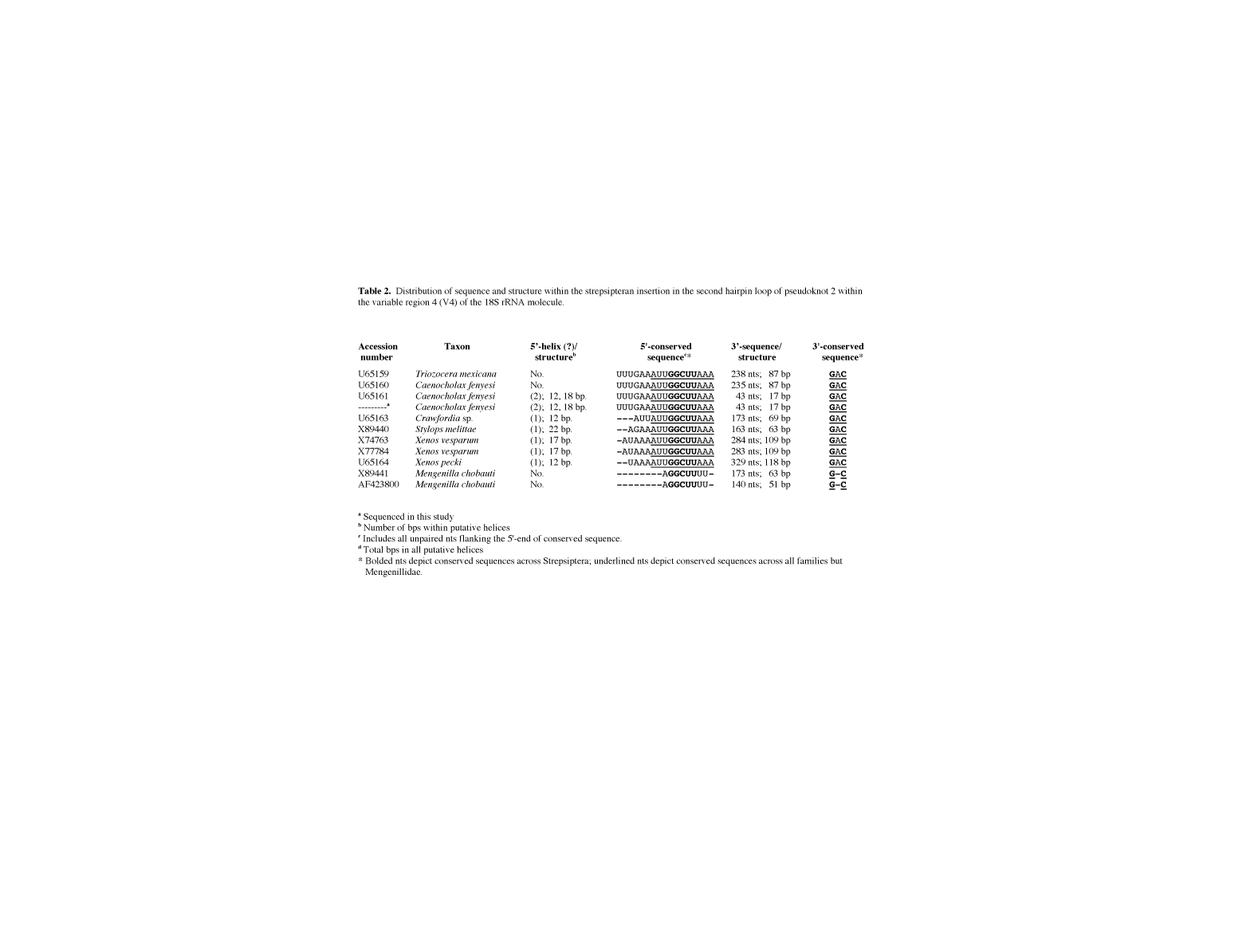
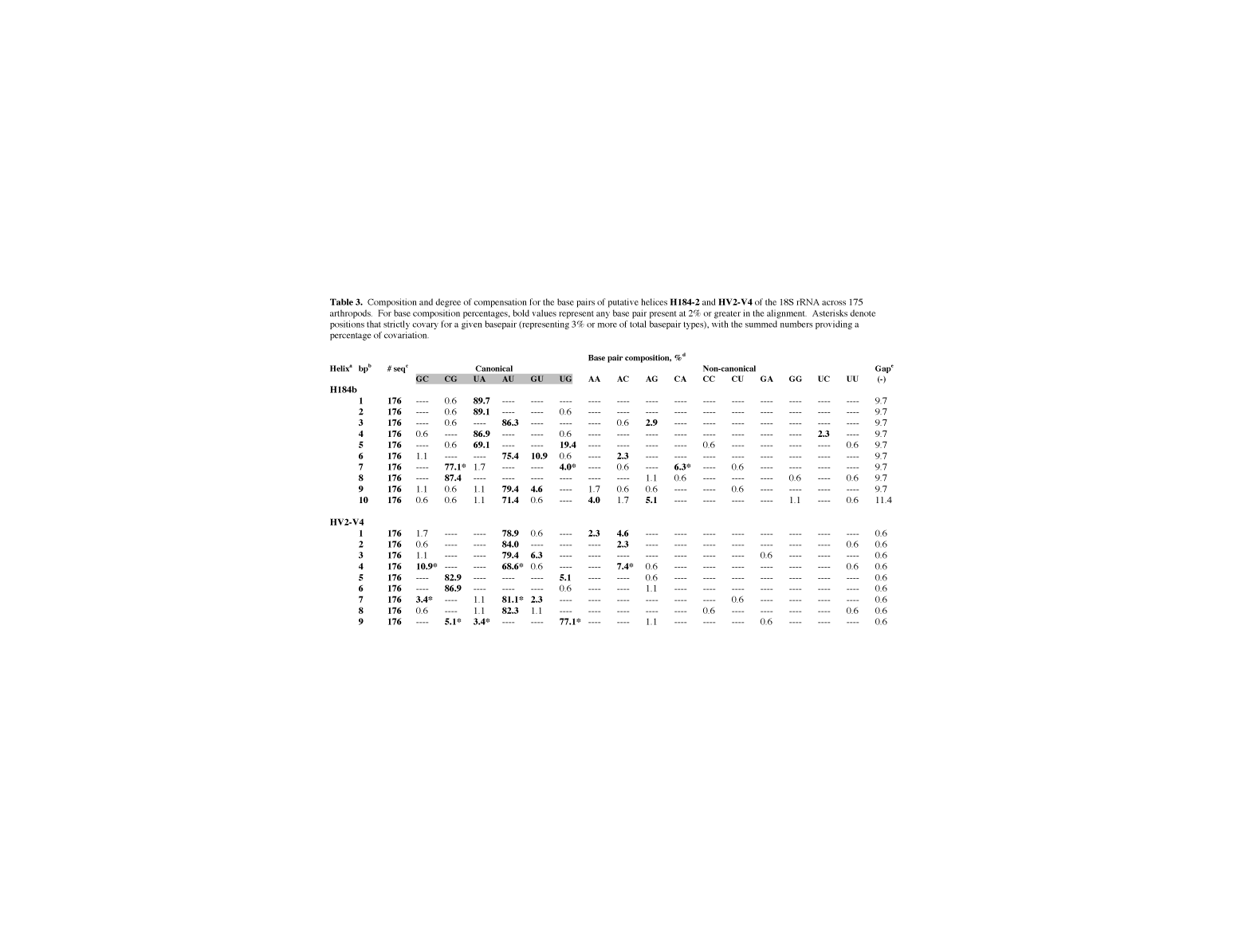
table 3